Proteomics BIOS E40 Home Assignment #3
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
Proteomics BIOS E40 Home Assignment #3
Due on Tuesday November 14 2023 (course website)
Part I (Undergraduate and Graduate) 22 pts
Recently, scientists have demonstrated that several chemical reagents can be used for the selective cleavage of peptide bonds in proteins: for example, S-ethyl trifluorothioacetate (s-
ETA) selectively cleaves the peptide bond located on the amino-side of Ser and Thr residues, and similarly, pentafluoropropionic acid (PFPA) selectively cleaves the peptide bonds located on the carboxyl-side of Asp residues .
Question 1:
The a-melanocyte stimulating hormone is a short peptide whose sequence is:
NH2-SYSMEHFRWGKPV-COOH
The peptide was submitted to chemical cleavage with s-ETA, and product of cleavage was sequenced by Edman degradation
1-a) 4pts. Predict the sequence of all the peptides formed by chemical cleavage.
1-b) 4 pts The HPLC profiles (Figure 1) show the derivatized amino acid produced after round 1, 2, and 3 of Edman degradation. Is your previous prediction confirmed by theses profiles? Briefly explain .
Question 2:
The heat shock protein tetradecapeptide has the following sequence:
NH2-CLNRQLSSGVSEIR-COOH
This protein was treated with s-ETA and the cleavage fragments were analyzed by MALDI.
2-a) 4 pts Predict the sequence of all the cleaved peptides (assuming complete cleavage) .
2-b) 4 pts The mass spectrum (Figure 2) shows that only 4 peaks were produced. Identify the
sequence of these 4 peptides using their measured mass and the table of masses that you got in one of your lecture handouts. Note: One peak is produced by a post-translationally modified form of the uncleaved peptide.
Question 3:
The heat shock tetradecapeptide was also treated with PFFA, and 30% of the original peptide (Mass: 1642.8 Da) was converted into a smaller peptide (mass: 1562.8 Da) .
3-a) 2 pts Did you expect any cleavage with PFFA? Briefly explain .
3-b) 2 pts Provide an explanation for this observation using the mass difference between the original and the smaller peptide and from what you have learned in class about protein phosphorylation .
3-c) 2 pts Suggest one method to confirm your hypothesis .
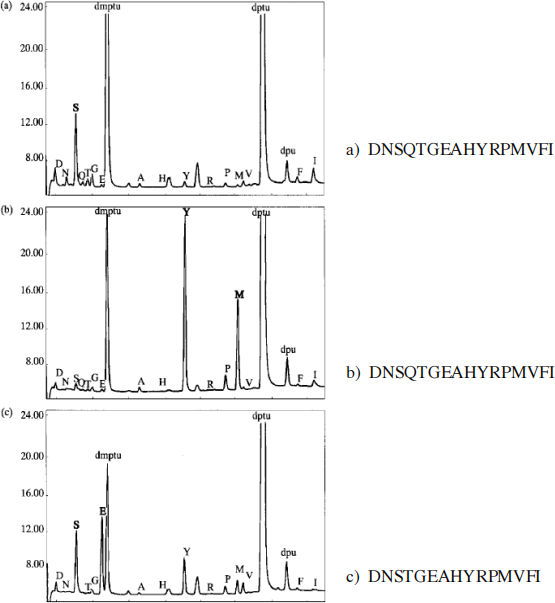
FIGURE 1: HPLC profiles after Edman degradation. (see question 1b). Panels a, b, and c represent the profile after 1, 2, and 3 rounds of Edman degradation, respectively. If you cannot read the labels placed on the top of each AA- PTH peaks on the profiles please refer to the series of letters placed on the right side of each panel. These letters correspond to peak labels (from left to right). The 2 very large peaks corresponding to the Edman reaction by-products dmptu and dptu are not included in the series of labels .
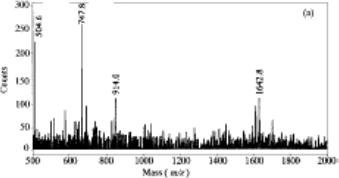
FIGURE 2:
Mass spectrum representing the masses of peptides produced by the treatment of the heat shock protein tetradecapeptide with s-ETA (See question 2b). The m/z ratio for the 4 labeled peaks are: m/z = 504.6, 747.8, 914.0, 1642.8
Part II (Undergraduate and Graduate) 18 pts
Purified polysaccharides were submitted to the following analysis:
Analysis #1: The polysaccharide was submitted to acid hydrolysis to break to glycosidic bonds and the products of the reaction were analyzed by mass spectrometry. The mass spectrum shows a major peak corresponding to a 180 Da residue. This residue was identified as glucose. A minor peak corresponding to a 342 Da residue was also observed. Note: The following masses may be useful: C: 12 Da; O: 16 Da; and H: 1Da

ANSWER THE FOLLOWING QUESTIONS
a) 2 pts Is the glucose molecule diagramed above an a or a β anomer? Briefly explain .
b) 4 pts What is the identity of the compound leading to the minor peak and why is the mass of this compound 342 Da (show your calculation)?
Analysis #2: The diagram below shows a linear polysaccharide characterized by a (1à4)
glycosidic linkage and containing only glucose residues. This oligosaccharide was subjected to permethylation, a reaction replacing the hydrogen of a hydroxyl group by CH;(R-OH + ICH3 à R-OCH3 + IH)
The permethylated polysaccharide was then submitted to acid hydrolysis .
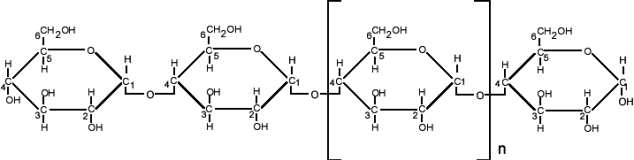
ANSWER THE FOLLOWING QUESTION
c) 2 pts Among the three modified glucose residues listed below which one should be the most abundant?
2,3,6-tri-O-methyl-glucose; 1,2,3,6-tetra-O-methyl glucose; 2, 3, 4, 6-tetra-O-methyl glucose . (Note that the numbers refer to the carbons carrying the methyl groups)
Analysis #3: A branched polysaccharide containing only glucose residues was subjected to the treatments described in analysis 2. The analysis of the products revealed three methylated sugars in the ratio 20/1/ 1: The methylated sugars were 2,3,4-tri-O-methyl glucose (20), 2,4-di-O-methyl glucose (1), 2,3,4,6-tetra-O-methyl glucose (1) .
ANSWER THE FOLLOWING QUESTIONS
d) 4 pts. Based on the ratio of these modified sugars, what is the predominant glycosidic linkage between the glucose residues? Explain .
e) 2 pts. Why do the branch points occur through the carbon C-3?
f) 4 pts. In the sentence, “In this polysaccharide a branch occurs at a frequency of one every X residue” replace X by a probable numerical value. Briefly explain .
X =
Part III (Graduate Students Only) 40 pts (+ 4 bonus)
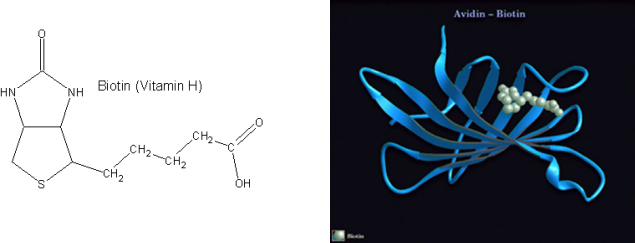
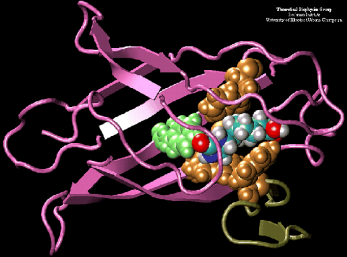
Avidin is a small, basic glycoprotein found in the egg white and tissue of birds, reptiles, and amphibians that binds to the small molecule biotin (biotin is also known as vitamin H, see structure below). The avidin/biotin complex is the strongest known noncovalent biological interaction between a protein and ligand, with an association constant (Ka) of 1015 M- 1. Analysis of the avidin crystal structure reveals a highly non-polar “pocket” in which the biotin molecule binds (crystal structure at right); the nature of the interaction is highlighted (crystal structure at bottom) when the hydrophobic amino acids (gold and green) that line the pocket and interact with the non-polar biotin molecule (blue, red and grey) are represented as a space-filling model.
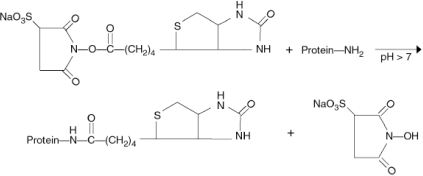
The high affinity and specificity of avidin-biotin interactions have been exploited for diverse applications in immunology, histochemistry, in situ hybridization, affinity chromatography and many other areas. Many years ago, it was shown that N-hydroxysuccinimide (NHS) esters of biotin could be used to covalently attach biotin to proteins, a process commonly referred to as “biotinylation”. Since the target for the NHS ester is the deprotenated form of the primary amine, the reaction of NHS esters with proteins at neutral pH (or higher) is favorable. The reaction of a commercially available biotinylation reagent, Sulfo- NHS-Biotin, with primary amines is shown below. Once tagged with biotin, a molecule of interest can be used to probe cells, tissues or proteins mobilized on blots or arrays, and the tagged molecule can be detected with a labeled avidin conjugate.
Membrane proteins are often critical components of signaling pathways that regulate diverse biological responses, and are of substantial interest with regard to various aspects of disease, from molecular diagnosis to therapeutics. The development of membrane impermeable biotinylation reagents that can be used selectively biotinylate membrane proteins has provided researchers with a way to specifically detect and purify this important class of biomolecules, which are often present at low abundance compared to cellular proteins.
ANALYSIS #1
Recently, one group of researchers published a paper utilizing this technique. They
biotinylated plasma membrane proteins on living cells grown in vitro, lysed the cells, solubilized both the membrane and cellular proteins, and separated the solubilized proteins by 2D-PAGE.
The proteins were detected either by silver staining of the 2D PAGE gels (panel B), or by transferring the proteins from the 2D PAGE gels to PDVF membranes and hybridizing them with horse radish peroxidase (HRP) conjugated avidin (panel A). HRP conjugated avidin is a hybrid protein composed of avidin (which binds biotin) and HRP (which catalyzes a colorimetric – or color change – reaction), allowing for the detection of the avidin molecule by the HRP-catalyzed color change with sensitivity comparable to that of a western blot (immunoblot).
ANSWER THE FOLLOWING QUESTIONS
Question 1 (10 pts) : The authors claim that using biotinylation of surface proteins, they “are able to obtain a selective and enhanced visualization of low abundance proteins through
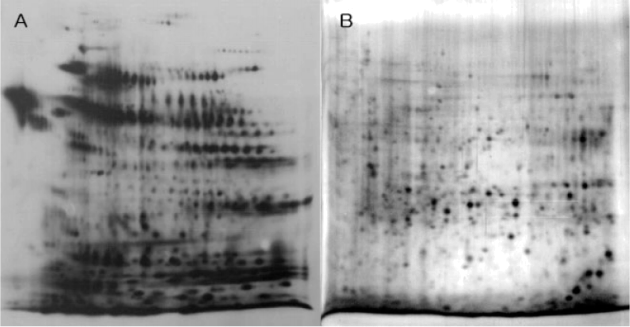
biotinylation of the surface membrane”. Examine panels A and B. Given the data presented, do you agree with their claim?
(note: the quality of the figure in this home assignment is identical to the quality of the figure provided by the authors in their published paper.)
Question 2 ( 5pts + 2 bonus): Based on what you have learned in lecture, explain the characteristic distribution pattern of biotinylated proteins in panel A: how can you explain the presence of rows of protein with the same molecular weight but different isoelectric points?
Question 3 (10 pts) : It is possible to purchase immobilized avidin beads, in which the avidin protein has been covalently linked to very small (but visible) beads. This product is often used for the purification of biotinylated proteins. Outline the experimental steps you would perform in order to purify membrane proteins, using the following reagents: cells, a membrane impermeable biotinylation reagent, immobilized avidin beads, and excess biotin.
Question 4 ( 5 pts): During the labeling experiment, a researcher mistakenly lysed the cells before adding the biotinylation reagent. Explain the consequence of the researcher’s mistake.
ANALYSIS #2
The authors referenced in question 1 purified their biotinylated proteins using immobilized avidin beads, and separated the purified proteins by 2D-PAGE. As in question 1, purified proteins were visualized by silver staining (RIGHT panel) and hybridization with HRP-conjugated avidin (LEFT panel).
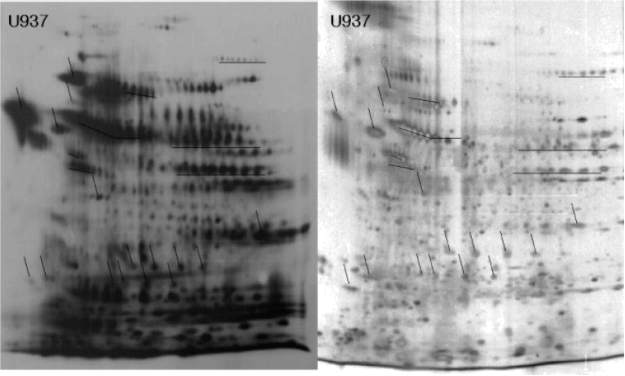
ANSWER THE FOLLOWING QUESTIONS
Question 5 (5 pts): Compare the above panels to the panels A and B from question 1. Do you think these authors have succeeded in at least partially purifying biotinylated proteins? Clearly describe the interpretation of the data that led you to your answer.
Question 6 (3 pts): Two spots from the silver-stained gel were isolated and submitted to in gel-trypsin digestion. The tryptic peptides were analyzed by MALDI-TOF and the monoisotopic masses of some of these peptides are listed in the table below.
|
Spot 1 (Mass range: 33-35 kDa) |
Spot 2 (Mass range 68-79 kDa) |
|
1785.876 |
1316.629 |
|
1465.791 |
1430.683 |
|
1342.686 |
1566.772 |
|
1175.551 |
1677.8 |
|
1062.521 |
1818.831 |
|
932.520 |
1887.963 |
Identify both proteins using MS-FIT (Protein Prospector). You should not have to change any of the default values to perform the search.
Question 7 (2 pts): For each result, write down the name of the protein identified by peptide mass fingerprinting, and whether or not you are confident in the results of the peptide mass fingerprinting search. If you read about each of the proteins by following the link to the SWISS-PROT entry, you’ll find something unexpected about at least one of these proteins.
What is it? (HINT: Think about how these proteins were purified.)
Question 8: (bonus question, 2 pts): Suggest a biological interpretation of the unexpected result.
2023-12-15