BIOS E-40- HA #2
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
BIOS E-40- HA #2
DUE DATE: October 17 2023
PART 1: Undergraduate + Graduate, (20 points)
A tryptic peptide was analyzed with a triple-quadrupole mass spectrometer in ms/ms mode. The following figure shows the mass spectrum after fragmentation (Panel A) and an enlarged detail of the spectrum (Panel B) .
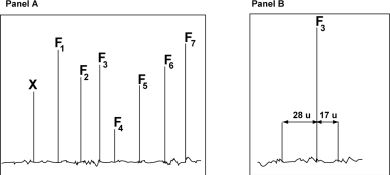
The following table shows the m/z ratio of the fragments F1 to F7 determined on the spectrum shown in Panel A.

a) Complete the table and identify the peaks that belong to the same series. The table showing the contributions of residues to the m/z ratio of a peptide can be found in your lecture notes. Briefly explain your answer (4 pts) .
b) Determine the partial sequence of the fragmented peptide. The trypsin digestion was complete and the fragmented peptide did not contain any missed cleavage site. (2 pts)
c) A careful analysis of the region of the spectrum indicates the presence of two satellites peak flanking F3 (see Panel B above). Determine the nature of the F3 fragment (y-ion or b-ion) based on the difference in m/z ratio between these satellite peaks and F3. Briefly explain. (4 pts)
You may need to following information to answer the question:
Atomic mass: Oxygen: 16; Nitrogen: 14; Hydrogen: 1; Carbon: 12
d) Based on your previous answer, write the sequence of tryptic peptide from its amino- end to its carboxy-end (2 pts)
e) Peak X (Panel A) is produced by a multi-protonated form of the ion responsible for the formation of the peak F2 (contain only 1 proton) . Calculate how many protons are associated with peak X knowing that the m/z ratio for this peak is: 160.08. Show your calculation (4 pts).
f) The fragmentation spectrum of a mutant form of the protein where the N-terminal residue of the F2 ion is replaced by alanine (A) shows a shift of peaks F2 and X.
Calculate the m/z shift of the peak X due to the mutation. Show your calculation (4 pts).
PART 2: Graduate only (30 points)
In proteins disulfide bonds are formed by the oxidative linkage of sulfhydryl groups (-SH) carried by the side chain of cysteine residues .
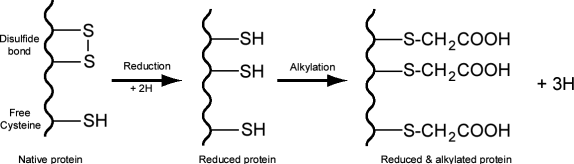
Mass spectrometer can detect a change in mass due to reduction and alkylation of disulfide bonds or free sulfhydryl groups. In the rest of this question we will use the following terminology:
Mnat = mass of the native protein
Mr = mass of the reduced protein
Mr+a = mass of the reduced and alkylated protein
Ma = mass of the alkylated protein without reduction
a) Based on the atomic mass of H (1), C (12), O (16), S(32) calculate the change in protein mass due to the reduction and alkylation of one cysteine residue engaged in a disulfide bond and the change in mass due to the alkylation of a free cysteine residue. (8 pts)
b) Determine the equation linking Mr+a , Mnat , NSS (total number of cysteine residues engaged in disulfide bonds) and NSH (total number of free cysteine). (6 pts)
c) Determine the equation linking Ma , Mnat and NSH . (6 pts)
d) By combining the two previous equations calculate the number of disulfide bonds contained in a protein that has the following characteristics: Mnat = 2000 Da, Mr+a = 2586 Da and Ma = 2232 Da. Show your calculation. (6 pts)
e) In which region(s) of the cell could you find a protein with a disulfide bond? Briefly explain (4 pts)
2023-12-14