BIOCHEMISTRY 3381A Assignment #2
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
BIOCHEMISTRY 3381A
Assignment #2 – Due Nov 6 in OWL & Gradescope
1. The following primary amino acid sequence is from a protein containing 345 residues. The N-terminal domain is highlighted in cyan (105), and the C-terminal domain is highlighted in yellow (100 residues).
MDPKRIRDGYIVKRGSVFQTWKPMWVLVLEDGVEFYKRKSDNSPKGMIPMKGSTLTSPCNDFGKRMFVFKVTTTKQNDHFFQAAFLDEREAWVRDIKRAIKCIEGFGGQKFARKSTRRSIRLPESIDMGALYLSLKDTEKGIKDLNLEKDKRIFNHCFTGNCVIEWLVSNQSVKNRQEGLMVASSLMNEGYLQPAGGDLSKSAIDGSAENPFLDQPDAFFYFPDSGGFFCEDNSSSEDDVILKEDGSGVSMQACLELQGHRGQWKVKFDLRGDPAYLHFYAPDDGCVYITVYGSGITVCASAGSEGVCIYIYGSTGVHFYLQAGTPKALREWARKAIQKAMRTGK
a) Based on your knowledge of factors contributing to different types of secondary structure, predict what secondary structure elements (and connections) are likely to be formed for the C-terminal domain (highlighted in yellow)? Use a table to summarize your predictions. What type(s) of motifs and domain will this arrangement of secondary structure elements form for the C-terminal region of the protein (Hint: the domain is listed in your notes in lecture 6, but is not one we discussed in detail)? Include a figure to illustrate motif(s) and domain. Justify the basis for making all predictions. (max length is 1-page 1.15 spacing including tables and figures). (15 marks)
b) What is the net charge of the yellow domain at pH = 1, 7.5 and 14 (assuming full de/protonation if pH is above or below pKa). Remember to include the C-terminal carboxylate group in your calculations. Show a table of amino acids and charges to justify your answer. (3 marks)
c) What kind of adsorption chromatography column would you expect this 345 residue protein to bind at pH = 7.0 under (a) low salt conditions (ie. 100 mM NaCl), and (b) high salt conditions (ie. 2.5 M ammonium sulfate)? Use Expasy ProtParam to calculate the pI of the protein. Explain both column choices using the principle of salting in/out? (max 500 words) (6 marks)
d) This protein is from a species of hominid identified in the 1930’s called homo longi (aka Dragon Man) and appears to be structurally and (perhaps) functionally related to an important protein from homo sapiens. Based on sequence similarity, identify what the new protein is and discuss possible function(s) that it might have? Where would you expect to find this protein in the cell. Given your prediction for function of the C-terminal domain and the identity of the human homolog, what function might the N-terminal domain (coloured in cyan) of the protein serve? Explain how you came to your conclusions. (max 300 words) (6 marks)
e) Download (from the protein data bank) the first x-ray crystal structure of the human homologue of the C-terminal domain of your protein in complex with a small molecule ligand (hint, this structure was determined in 2007 by the Junop lab). Create a figure (with legend) using Pymol that shows a zoomed in view of the ligand binding region (this should include the protein represented in cartoon; and the ligandand interacting side chains in stick representation). Use colour wisely to help show these interactions clearly to your audience. (7 marks)
f) Run a computational tertiary structure prediction (using Phyre2) for the C-terminal domain of your protein and align this predicted structure (using Pymol) with the structural homologue downloaded from the protein data bank in question e). Create a figure of the two aligned structures using Pymol and briefly discuss any significant differences observed between your prediction (in 1a), the computational prediction (f) and the homologue structure downloaded from the protein data bank. (8 marks) (max 300 words)
QUALITY OF PRESENTATION:
Grammar, clarity of logic/reasoning in writing, aesthetics (5 marks)
RUBRIC
1. A)
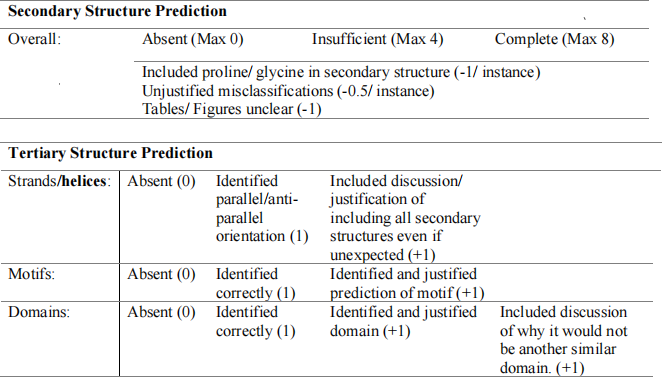
B) - One mark for correctly determining charge at each of pH.
C)
- Must have either calculated net charge or pI for entire protein at pH 7.0. No need to show work, just give a number and sentence or two on how it was calculated. (1 mark)
- (a) at low salt = (0.5 for getting column correct)
- (a) Explanation of salting in and how principle is used for chromatography (2 marks)
- (b) at high salt = (0.5 for getting column correct)
- (b) Explanation of salting out and how principle is used for (2 marks)
D)
- Correct ID of protein (1 mark)
- Explanation for how they ID the protein (can be any method as long as it makes sense) (1 mark)
- Explanation of where you would find protein in cell and why (1 mark)
- Correct ID of N-terminal domain function with short explanation for choice of domain (1 mark)
- Discussion of function – this will most likely come directly from literature. As long as the discussion mentions ligand and how the protein uses it (1 mark) and what that process is required for biologically (1 mark), give full marks.
E)
- Correct PDB file (1 mark)
- Figure components correct:
o Cartoon with only interacting residues in stick (1 mark)
o Properly zoomed in (and cropped if needed) so that the figure doesn’t contain a lot of extra detail that is not required for illustrating ligand and interacting residues (1 mark)
o Correct ligand (0.5 mark), and correct interacting residues (0.5 mark)
o Figure labelled so that each residue and ligand is clearly labelled (label NOT on top of residue) (1 mark). This will almost certainly require more than a single angle to show all residues clearly without overlapping. If they choose one figure, it must be very clear and easy to interpret data. (1 mark)
o Wise use of colour to contrast ligand, interacting residues and remainder of protein. Can be any combo of colours and effects (transparency, surface, etc) (1 mark)
o Figure legend with accurate title and description – all colours described, pdb code, etc. (1 mark)
F)
- Phyre2, I-tassar (or other method) to generate predicted pdb file of JUST yellow domain sequence, this should not have the N-term sequence included (0.5 mark)
- Correctly aligned phyre model with human homologue downloaded from PDB (0.5 mark)
- Figure components correct:
o cartoon representation, no sticks so alignment is easier to see (0.5 mark)
o Figure labelled so that each protein is clearly identified (label NOT on top of protein) (0.5 mark). Need to indicate, using label and/or colour, etc the regions on the figure that are different. This is important so they can refer to figure in explanation of differences in structure. May require more than one angle to show all differences between two structures. If they choose one figure, it must be very clear and easy to interpret. (1 mark)
o Wise use of colour to enhance contrast of two aligned protein structures (1 mark)
o Figure legend with accurate title and description – all colours described, pdb code, etc. (1 mark)
o Description of differences in models
§ Difference between their manual prediction from a) vs one from PDB
§ Difference between their manual prediction from a) vs Phyre model
§ Difference between their manual prediction Phyre model vs PDB model
§ Each worth 0.5 mark for identifying difference (or no diff if that is the case) and 0.5 mark for suggesting a reason difference might have occurred – only need to address differences involving changes in secondary structure elements.
QUALITY OF PRESENTATION:
This is an overall impression mark:
§ Good grammar (1 mark)
§ Proper referencing and in text citation (any style is ok) (1 mark)
§ Questions answered in clear, concise manner (2 mark)
§ Overall appearance of report. Well-spaced, centered, etc. (1 mark)
2023-10-31