ECS607U Data Mining Assignment 2
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
Assignment 2
Task 1
In this task we need to write a function to return Kulczynski measure. The Kulczynski measure is defined as:

The Groceries dataset is selected for itemsets, it contains 38765 rows for Market Basket Analysis. Some data samples are as follows:

In Kulczynski funciton, we need to calculate the support for A, B, A and B. The function code is as follows:
import pandas aspd
import numpy as np
def kulczynski(iterm_sets, rule):
rule a part, rule b part = rule
support a = sum(groceries_data['itemDescription![]() apply(
apply(
lambda x : len(set(x)&set(rule a part)
)== len(rule a part)
support b= sum(groceries_data['itemDescription![]() apply(
apply(
lambda x : len(set(x)&set(rule b part)
)== len(rule b part)
support a and b= sum(groceries_data['itemDescription![]() apply(
apply(
lambda x : len(set(x)&set(rule a part+ rule b part)
)== len(rule a part+ rule b part)
return(support a and b/support a + support a and b/support b)*
0.5
Task 2
In this task we need to write a function to return imbalance ratio. The imbalance ratio is defined as:
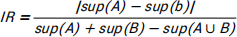
The Groceries dataset is selected for itemsets. In imbalance ratio funciton, we need to calculate the support for A, B, A and B. The function code is as follows:
import pandas aspd
import numpy as np
def imbalance_ratio(iterm_sets, rule):
rule a part, rule b part = rule
support a = sum(groceries_data['itemDescription![]() apply(
apply(
lambda x : len(set(x)&set(rule a part)
)== len(rule a part)
support b= sum(groceries_data['itemDescription![]() apply(
apply(
lambda x : len(set(x)&set(rule b part)
)== len(rule b part)
support a and b= sum(groceries_data['itemDescription![]() apply(
apply(
lambda x : len(set(x)&set(rule a part+ rule b part) )== len(rule a part+ rule b part)
return np.abs(support a - support b) / (support a + support b - support a and b)
Task 3
Excluding the case of a single item, there contiains (N-1)! possible valid itemsets.
Task 4
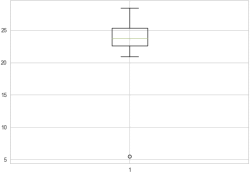
Through the box plot we can determine the outlier, the outlier is 5.49. The boxplot regards outliers as points in the outer 1% of a normal population.
Task 5
After reading the dataset, the percentage of changes was calculated, and a one-class SVM classifier was used to identify outliers. The 3D scatterplot of the dataset with object is color-coded outlier is shown in the figure below.
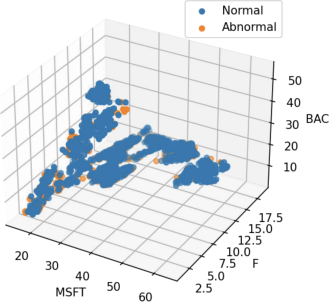
The histogram and the frequencies of the abnormal detection results are shown in the figure below, in which 18% of the samples are abnormal samples. The results of One-class SVM are not based on distance and density, from distance.

import pandas aspd
#Load CSVfile, setthe 'Date'values asthe index ofeach row, anddisplay the first rows ofthe dataframe
stocks = pd.read excel('stocks.xlsx header='infer')
stocks.index = stocks['Date]'
stocks = stocks.drop(['Date![]() axis=1)
axis=1)
N,d= stocks.shape
delta = pd.DataFrame(
100*np.divide(stocks.iloc[1:,:].values-stocks.iloc[:N-1,:].values, stocks.iloc[:N-1,:].values),
columns=stocks.columns, index=stocks.iloc[1:].index
)
delta.head()
from sklearn.svm import OneClassSVM
ee = OneClassSVM(nu=0.01, gamma='auto')
yhat = ee.fit predict(delta)
import matplotlib.pyplot asplt
fig = plt.figure(dpi=150)
ax = fig.add subplot(projection='3d')
ax.scater(stocks['MSFT![]() iloc[np.where(yhat==1)[0]+ 1],
iloc[np.where(yhat==1)[0]+ 1],
stocks['F![]() iloc[np.where(yhat==1)[0]+ 1],
iloc[np.where(yhat==1)[0]+ 1],
stocks['BAC![]() iloc[np.where(yhat==1)[0]+ 1])
iloc[np.where(yhat==1)[0]+ 1])
ax.scater(stocks['MSFT![]() iloc[np.where(yhat==-1)[0]+ 1],
iloc[np.where(yhat==-1)[0]+ 1],
stocks['F![]() iloc[np.where(yhat==-1)[0]+ 1],
iloc[np.where(yhat==-1)[0]+ 1],
stocks['BAC![]() iloc[np.where(yhat==-1)[0]+ 1])
iloc[np.where(yhat==-1)[0]+ 1])
plt.legend(['Normal 'Abnormal]')
ax.set xlabel('MSFT')
ax.set ylabel('F')
ax.set zlabel('BAC')
Task 6
The data set is first subjected to PCA dimensionality reduction, and then the distance of each sample is obtained by nearest neighbor calculation. We plotted the sample distances as boxplots. As shown in the figure below, samples with a sample distance greater than 2.5 can be regarded as outliers.

The scatter diagram of PCA is shown in the figure below. It can be seen that the distance between the outlier point and other points is relatively long.

df= pd.read csv(url, header=None)
data = df.values
X, y = data[:, :-1], data[:, -1]
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
X pca = pca.fittransform(X)
from sklearn.neighbors import NearestNeighbors
knn = NearestNeighbors(n_neighbors=2)
knn.fit(X pca)
plt.scater(X pca[distances.mean(1)<2.5, 0], X pca[distances.mean(1)<2.5, 1])
plt.scater(X pca[distances.mean(1)>2.5, 0], X pca[distances.mean(1)>2.5, 1])
plt.legend(['Normal 'Abnormal]')
Task 7
There are many HTML tags in the web page, such as html, body, h1, p, table, thead, tr, th, rd. By consulting the introduction of the HTML web page, the meaning of each tag is as follows:
. html, page start and end
. body, content start and end
. h1, header
. p, paragraph
. table, table start and end
. thead, header of table
. tr, row of table
. th, head row of table
. td, content row of table
With Beautiful Soup, data is crawled and stored in tables. The result is shown in the figure below.

from bs4import BeautifulSoup
data =
BeautifulSoup(requests.get('htp:/eecs.qmul.ac.uk/~emmanouilb/income _table.html![]() text)
text)
header = [x.text.strip()for x in table.find al(t'head')[0].find al(t'h')]
table = data.find(t'able')
table_body = table.find(t'body')
rows = table_body.find al(t'r')
table_data = []
for row in rows:
cols = row.find al(t'd')
cols = [ele.text.strip()for ele in cols]
table_data.append([ele for ele in cols ifele])
table_data = pd.DataFrame(table_data, columns=header)
Task 8
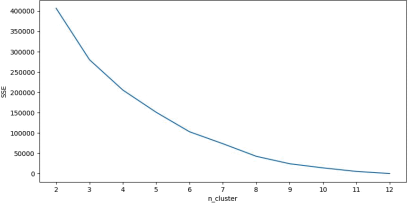
Through Beautiful Soup, we crawl webpages with multiple keywords. And the frequency of words in the article is counted, and then the web pages are clustered. We experimented with various numbers of clusters and recorded the clustering loss. The elbow diagram is shown in the figure below. It can be seen from the figure that the optimal number of clusters is 6.
The final clustering results are as follows. The clustering results are more reasonable. Semi-supervised and supervised learning are in the same
category, and data mining and data warehouse are in the same category.
1. ['unsupervised learning' 'anomaly detection' 'dimensionality reduction' 'statistical classification']
2. ['association rule learning']
3. ['data mining' 'data warehouse']
4. ['supervised learning' 'semi-supervised learning' 'online analytical processing']
5. ['cluster analysis']
6. ['regression analysis']
data_texts = []
for keyword in keywords:
keyword= keyword[0].upper()+keyword[1:]
keyword= keyword.replace(' ' ')
data = requests.get('htps:/en.wikipedia.org/wiki/' + keyword).text data_text= BeautifulSoup(data).find(d'iv atrs={'class':
'mw-parser-output}').text
data_texts.append(data_text.lower()
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.cluster import Kmeans
data_bag_of words =
CountVectorizer(max_features=200).fittransform(data_texts)
sse = []
for n_clusters in range(2, 13):
km = KMeans(n_clusters=n_clusters)
km.fit(data_bag_of words)
sse.append(km.inertia_)
km = KMeans(n_clusters=6)
km.fit(data_bag_of words)
for idx in range(6):
print(np.array(keywords)[km.labels_ == idx])
2023-08-04